 | Comparative transcriptomics reveals human-specific cortical features |  |
INTRODUCTION The cerebral cortex is involved in complex cognitive functions such as language. Although the diversity and organization of cortical cell types has been extensively studied in several mammalian species, human cortical specializations that may underlie our distinctive cognitive abilities remain poorly understood.
RATIONALE Single-nucleus RNA sequencing (snRNA-seq) offers a relatively unbiased characterization of cellular diversity of brain regions. Comparative transcriptomic analysis enables the identification of molecular and cellular features that are conserved and specialized but is often limited by the number of species analyzed. We applied deep transcriptomic profiling of the cerebral cortex of humans and four nonhuman primate (NHP) species to identify homologous cell types and human specializations.
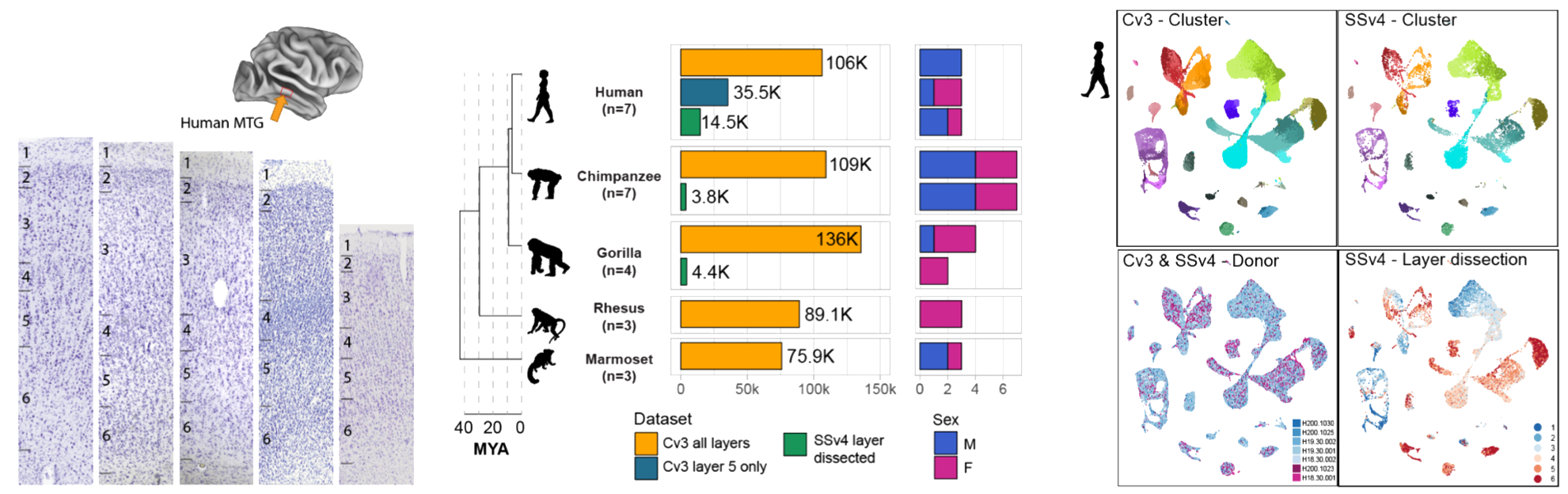
RESULTS We generated snRNA-seq data from humans, chimpanzees, gorillas, rhesus macaques, and marmosets (more than 570,000 nuclei in total) to build a cellular classification of a language-associated region of the cortex, the middle temporal gyrus (MTG), in each species and a consensus primate taxonomy. Cell-type proportions and distributions across cortical layers are highly conserved among great apes, whereas marmosets have higher proportions of L5/6 IT CAR3 and L5 ET excitatory neurons and Chandelier inhibitory neurons. This strongly points to the possibility that other cellular features drive human-specific cortical evolution. Profiling gorillas enabled discrimination of which human and chimpanzee expression differences are specialized in humans. We discovered that chimpanzee neurons have gene expression profiles that are more similar to those of gorilla neurons than to those of human neurons, despite chimpanzees and humans sharing a more-recent common ancestor. By contrast, glial expression changes were consistent with evolutionary distances and were more rapid than neuronal expression changes in all species. Thus, our data support a faster divergence of neuronal, but not glial, expression on the human lineage. For all primate species, many differentially expressed genes (DEGs) were specific to one or a few cell types and were significantly enriched in molecular pathways related to synaptic connectivity and signaling. Hundreds of genes had human-specific differences in transcript isoform usage, and these genes were largely distinct from DEGs. We leveraged published datasets to link human-specific DEGs to regions of the genome with human-accelerated mutations or deletions (HARs and hCONDELs). This led to the surprising discovery that a large fraction of human-specific DEGs (15 to 40%), and particularly those associated with synaptic connections and signaling, were near these genomic regions that are under adaptive selection.
CONCLUSION Our study found that MTG cell types are largely conserved across approximately 40 million years of primate evolution, and the composition and spatial positioning of cell types are shared among great apes. In each species, hundreds of genes exhibit cell type-specific expression changes, particularly in pathways related to neuronal and glial communication. Human-specific DEGs are enriched near likely adaptive genomic changes and are poised to contribute to human-specialized cortical function.
Images and movies
BibTex references
@Article { JSESCKYCLSTBBBCDEGGKMPRSSTTFHHKLMLSSWDGLHB23,
author = "Jorstad, Nicholas L. and Song, Janet H. T. and Exposito-Alonso, David and Suresh, Hamsini and Castro-Pacheco,
Nathan and Krienen, Fenna M. and Yanny, Anna Marie and Close, Jennie and Long, Brian and Seeman, Stephanie C.
and Travaglini, Kyle J. and Basu, Soumyadeep and Beaudin, Marc and Bertagnolli, Darren and Crow, Megan and
Ding, Song-Lin and Eggermont, Jeroen and Glandon, Alexandra and Goldy, Jeff and Kroes, Thomas and McMillen,
Delissa and Pham, Thanh and Rimorin, Christine and Siletti, Kimberly and Somasundaram, Saroja and Tieu,
Michael and Torkelson, Amy and Feng, Guoping and Hopkins, Guoping and H\öllt, Thomas and Keene, Dirk and
Linnarsson, Sten and McCarroll, Steven A. and Lelieveldt, Boudewijn P.F. and Sherwood, Chet C. and Smith,
Kimberly and Walsh, Christopher A. and Dobin, Alexander and Gillis, Jesse and Lein, Ed and Hodge, Rebecca and
Bakken, Trygve",
title = "Comparative transcriptomics reveals human-specific cortical features",
journal = "Science",
volume = "382",
pages = "eade9516",
year = "2023",
doi = "10.1126/science.ade9516",
url = "http://graphics.tudelft.nl/Publications-new/2023/JSESCKYCLSTBBBCDEGGKMPRSSTTFHHKLMLSSWDGLHB23"
}
![science.ade9516 summary.pdf [345Ko]](/Publications-new/images/pdf.png)


