 | Cytosplore-Transcriptomics: a Scalable Interactive Framework for Single-Cell RNA Sequencing Data Analysis |  |
bioRxiv - 2020
Download the publication :
![2020_cyto_transcriptomics.pdf [1.6Mo]](/Publications-new/images/pdf.png)
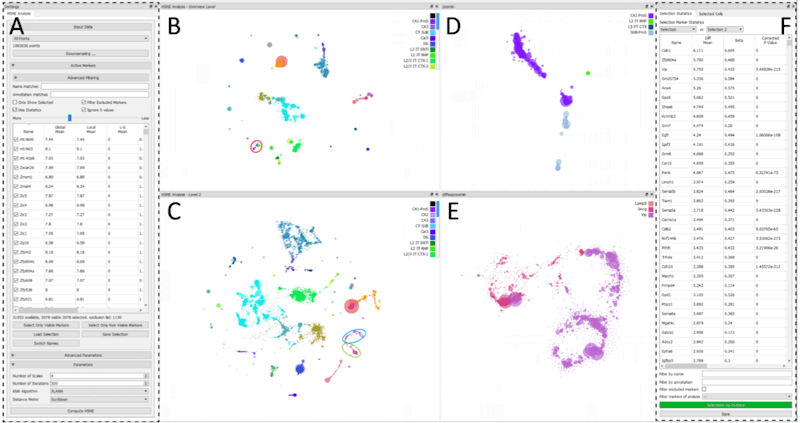
The ever-increasing number of analyzed cells in Single-cell RNA sequencing (scRNA-seq) experiments imposes several challenges on the data analysis. Current analysis methods lack scalability to large datasets hampering interactive visual exploration of the data. We present Cytosplore-Transcriptomics, a framework to analyze scRNA-seq data, including data preprocessing, visualization and downstream analysis. At its core, it uses a hierarchical, manifold preserving representation of the data that allows the inspection and annotation of scRNA-seq data at different levels of detail. Consequently, Cytosplore-Transcriptomics provides interactive analysis of the data using low-dimensional visualizations that scales to millions of cells.
Images and movies
|
| Other publications in the database
|
BibTex references
@Article { AEHMRL20,
author = "Abdelaal, Tamim and Eggermont, Jeroen and H\öllt, Thomas and Mahfouz, Ahmed and Reinders, Marcel and
Lelieveldt, Boudewijn P.F.",
title = "Cytosplore-Transcriptomics: a Scalable Interactive Framework for Single-Cell RNA Sequencing Data Analysis",
journal = "bioRxiv",
year = "2020",
doi = "10.1101/2020.12.11.421883",
url = "http://graphics.tudelft.nl/Publications-new/2020/AEHMRL20"
}
Back


![2020_cyto_transcriptomics.pdf [1.6Mo]](/Publications-new/images/pdf.png)
